#include "hawaii.h"#include "arena.h"#include "trajectory.h"#include "hep_hawaii.h"#include "angles_handler.hpp"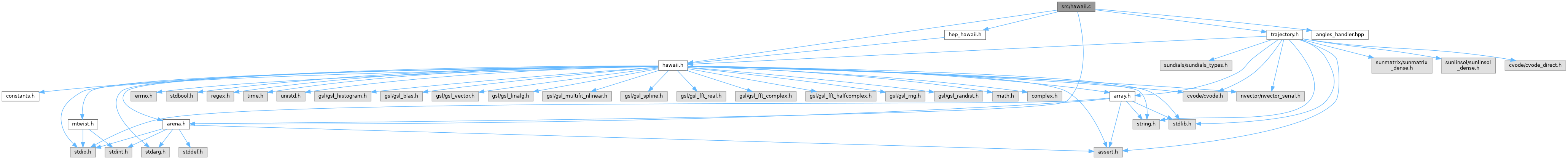
Macros | |
| #define | ARENA_REGION_DEFAULT_CAPACITY (8*1024) |
| #define | HISTOGRAM_MAX_TPS 50 |
| #define | R_HISTOGRAM_BINS 100 |
| #define | R_HISTOGRAM_MAX 10000000.0 |
| #define | DEFAULT_JINI_HISTOGRAM_FILENAME1 "jini.dat.1" |
| #define | DEFAULT_JINI_HISTOGRAM_FILENAME2 "jini.dat.2" |
| #define | DEFAULT_JINI_HISTOGRAM_BINS 352 |
| #define | DEFAULT_JINI_HISTOGRAM_MAX 35.0 |
| #define | DEFAULT_JFIN_HISTOGRAM_FILENAME1 "jfin.dat.1" |
| #define | DEFAULT_JFIN_HISTOGRAM_FILENAME2 "jfin.dat.2" |
| #define | DEFAULT_JFIN_HISTOGRAM_BINS 352 |
| #define | DEFAULT_JFIN_HISTOGRAM_MAX 35.0 |
| #define | DEFAULT_NSWITCH_HISTOGRAM_FILENAME1 "nswitch.dat.1" |
| #define | DEFAULT_NSWITCH_HISTOGRAM_FILENAME2 "nswitch.dat.2" |
| #define | DEFAULT_NSWITCH_HISTOGRAM_BINS 20 |
| #define | DEFAULT_NSWITCH_HISTOGRAM_MAX 20.0 |
| #define | ARENA_IMPLEMENTATION |
| #define | nparams 3 |
| #define | REAL(z, i) |
| #define | IMAG(z, i) |
Functions | |
| MoleculeSystem | init_ms_from_monomers (double mu, Monomer *m1, Monomer *m2, size_t seed) |
| Function init_ms_from_monomers prepares the MoleculeSystem based on the provided Monomer structs, sets up memory buffers and initializes random number generator. | |
| MoleculeSystem | init_ms (double mu, MonomerType t1, MonomerType t2, double *II1, double *II2, size_t seed) |
| Function init_ms prepares the MoleculeSystem based on the specified monomer types, sets up memory buffers and initializes random number generator. | |
| void | free_stored_histogram (StoredHistogram *sh) |
| void | free_ms (MoleculeSystem *ms) |
| const char * | display_monomer_type (MonomerType t) |
| double | find_closest_integer (double j) |
| find_closest_integer | |
| double | find_closest_half_integer (double j) |
| find_closest_half_integer | |
| void | j_monomer (Monomer *m, double j[3]) |
| j_monomer computes the magnitude of angular momentum of passed-in monomer. Currently, implemented only for linear molecules. | |
| double | torque_monomer (Monomer *m) |
| torque_monomer computes the magnitude of torque (time-derivative of angular momentum) of passed-in monomer. Requires the derivative of potential Monomer::dVdq to be set within the monomer. Currently, implemented only for linear molecules. | |
| void | rhsMonomer (Monomer *m, double *deriv) |
| rhsMonomer dispatches between computing the right-hand side of Hamilton's equations of motion depending on the type of monomer. In addition to differentiating the kinetic energy, the derivatives of potential energy, which are taken from Monomer::dVdq, are also added to compute the right-hand side. rhsMonomer does not account for requantization which should happen after this function has been called. | |
| void | extract_dVdq_and_write_into_monomers (MoleculeSystem *ms) |
| int | rhs (realtype t, N_Vector y, N_Vector ydot, void *data) |
| rhs function is passed to CVode library to propagate the trajectory. First, the phase-point coordinates are stored into MoleculeSystem struct. A contiguous vector of coordinates is assembled via extract_q_and_write_into_ms. Next, by calling the external function dpes, the derivatives of potential energy are computed and stored into MoleculeSystem::dVdq field. The components of derivative vector are then copied into the field Monomer::dVdq of the corresponding Monomer via the call to extract_dVdq_and_write_into_monomers. The right-hand side of Hamilton's equations with respect to intermolecular degrees of freedom are readily obtained and filled into output ydot, while the derivatives with respect to monomer's coordinates are handled by rhsMonomer function. | |
| gsl_matrix * | compute_numerical_jac (void(*transform_angles)(double *qlab, double *qmol), double *qlab, size_t ninput_coordinates, size_t noutput_coordinates, size_t order) |
| compute_numerical_jac | |
| Array | compute_numerical_derivatives (double(*f)(double *q), double *q, size_t len, size_t order) |
| compute_numerical_derivatives | |
| Array | compute_numerical_rhs (MoleculeSystem *ms, size_t order) |
| compute_numerical_rhs Computes the right-hand side of Hamilton's equations of motion using finite-difference formulas. The returned Array contains derivatives whose order corresponds to variable ordering employed in the MoleculeSystem structure. | |
| double | kinetic_energy (MoleculeSystem *ms) |
| Function kinetic_energy computes the kinetic energy at the phase-point stored in MoleculeSystem. | |
| void | put_qp_into_ms (MoleculeSystem *ms, Array qp) |
| void | get_qp_from_ms (MoleculeSystem *ms, Array *qp) |
| void | extract_q (double *qp, double *q, size_t QP_SIZE) |
| void | extract_q_and_write_into_ms (MoleculeSystem *ms) |
| void | invert_momenta (MoleculeSystem *ms) |
| invert_momenta | |
| double | Hamiltonian (MoleculeSystem *ms) |
| Function Hamiltonian calls kinetic_energy function to compute kinetic energy, assembles a contiguous vector of coordinates via calling extract_q_and_write_into_ms and passes it to external pes function. The obtained values are added and returned. | |
| double | generate_normal (double sigma) |
| void | q_generator (MoleculeSystem *ms, CalcParams *params) |
| Function q_generator generates | |
| void | p_generator_flux (MoleculeSystem *ms, double Temperature) |
| Function p_generator samples momenta | |
| void | p_generator (MoleculeSystem *ms, double Temperature) |
| Function p_generator samples momenta | |
| bool | reject (MoleculeSystem *ms, double Temperature, double pesmin) |
| reject applies the rejection step to the phase-point that is stored in the MoleculeSystem. It presupposes that the provided phase-point is sampled from | |
| void | calculate_M0 (MoleculeSystem *ms, CalcParams *params, double Temperature, double *m, double *q) |
| calculate_M0 Running mean/variance formulas taken from GSL 1.15 https://github.com/ampl/gsl/blob/master/monte/plain.c | |
| void | compute_dHdp (MoleculeSystem *ms, gsl_matrix *dHdp) |
| compute_dHdp | |
| void | calculate_M2 (MoleculeSystem *ms, CalcParams *params, double Temperature, double *m, double *q) |
| calculate_M2 Running mean/variance formulas taken from GSL 1.15 https://github.com/ampl/gsl/blob/master/monte/plain.c | |
| void | mpi_calculate_M0 (MoleculeSystem *ms, CalcParams *params, double Temperature, double *m, double *q) |
| mpi_calculate_M0 | |
| void | mpi_calculate_M2 (MoleculeSystem *ms, CalcParams *params, double Temperature, double *m, double *q) |
| mpi_calculate_M2 | |
| void | track_turning_points (Tracker *tr, double R) |
| int | correlation_eval_zimmerman_trick_free_metastable (Arena *a, MoleculeSystem *ms, Trajectory *traj, CalcParams *params, double *crln, size_t *tps, double Temperature) |
| correlation_eval_zimmerman_trick | |
| int | correlation_eval_zimmerman_trick (Arena *a, MoleculeSystem *ms, Trajectory *traj, CalcParams *params, double *crln, size_t *tps) |
| correlation_eval_zimmerman_trick | |
| void | check_if_requantization_should_be_applied_to_monomer (Monomer *m, size_t step_counter) |
| int | correlation_eval (Arena *a, MoleculeSystem *ms, Trajectory *traj, CalcParams *params, double *crln, size_t *tps) |
| correlation_eval | |
| CFncArray | calculate_correlation_array_and_save (MoleculeSystem *ms, CalcParams *params, double base_temperature) |
| calculate_correlation_array_and_save | |
| void | setup_nswitch_histogram_for_monomer (Monomer *m) |
| void | setup_jini_histogram_for_monomer (Monomer *m) |
| void | setup_jfin_histogram_for_monomer (Monomer *m) |
| void | send_histogram_and_reset (gsl_histogram *h) |
| void | normalize_cfnc (CFnc *cf) |
| CFnc | calculate_correlation_and_save (MoleculeSystem *ms, CalcParams *params, double Temperature) |
| calculate_correlation_and_save | |
| void | recv_histogram_and_append (Arena *a, int source, gsl_histogram **h) |
| void | gsl_fft_square (double *farr, size_t N) |
| SFnc | calculate_spectral_function_using_prmu_representation_and_save (MoleculeSystem *ms, CalcParams *params, double Temperature) |
| calculate_spectral_function_using_prmu_representation_and_save | |
| int | assert_float_is_equal_to (double estimate, double true_value, double abs_tolerance) |
| double * | linspace (double start, double end, size_t n) |
| double * | arena_linspace (Arena *a, double start, double end, size_t n) |
| size_t * | arena_linspace_size_t (Arena *a, size_t start, size_t end, size_t n) |
| bool | write_spectrum (const char *filename, Spectrum sp) |
| int | write_spectrum_ext (FILE *fp, Spectrum sp) |
| bool | write_correlation_function (const char *filename, CFnc cf) |
| int | write_correlation_function_ext (FILE *fp, CFnc cf) |
| bool | write_spectral_function (const char *filename, SFnc sf) |
| int | write_spectral_function_ext (FILE *fp, SFnc sf) |
| int | write_histogram_ext (FILE *fp, gsl_histogram *h, int count) |
| void | sb_reserve (String_Builder *sb, size_t n) |
| void | sb_append (String_Builder *sb, const char *line, size_t n) |
| void | sb_append_null (String_Builder *sb) |
| void | sb_reset (String_Builder *sb) |
| void | sb_append_cstring (String_Builder *sb, const char *line) |
| void | sb_append_format (String_Builder *sb, const char *format,...) |
| void | sb_append_seconds_as_datetime_string (String_Builder *sb, int s) |
| void | sb_free (String_Builder *sb) |
| bool | read_correlation_function (const char *filename, String_Builder *sb, CFnc *cf) |
| bool | average_correlation_functions (CFnc *average, CFncs cfncs) |
| int | average_correlation_functions__impl (CFnc *average, int arg_count,...) |
| bool | read_spectral_function (const char *filename, String_Builder *sb, SFnc *sf) |
| bool | parse_temperature_from_header (const char *header, double *Temperature) |
| bool | parse_number_of_trajectories_from_header (const char *header, double *ntraj) |
| bool | read_spectrum (const char *filename, String_Builder *sb, Spectrum *sp) |
| bool | writetxt (const char *filename, double *x, double *y, size_t len, const char *header) |
| double | analytic_full_partition_function_by_V (MoleculeSystem *ms, double Temperature) |
| gsl_histogram * | gsl_histogram_extend_right (gsl_histogram *h, size_t add_bins) |
| void | free_cfnc (CFnc cf) |
| void | free_cfnc_array (CFncArray ca) |
| void | free_sfnc (SFnc sf) |
| void | free_spectrum (Spectrum sp) |
| double | wingmodel (WingParams *wp, double t) |
| double | wingmodel_image (WingParams *wp, double nu) |
| int | wingmodel_f (const gsl_vector *x, void *data, gsl_vector *f) |
| int | wingmodel_df (const gsl_vector *x, void *data, gsl_matrix *J) |
| void | gsl_multifit_callback (const size_t iter, void *params, const gsl_multifit_nlinear_workspace *w) |
| void | gsl_nonlinear_opt (size_t n, double *x, double *y, WingParams *wing_params) |
| WingParams | fit_baseline (CFnc *cf, size_t EXT_RANGE_MIN) |
| void | kaiser_apodization (Array a, double sampling_time) |
| void | blackman_apodization (Array a, double sampling_time) |
| void | connes_apodization (Array a, double sampling_time) |
| double * | dct (double *v, size_t len) |
| double * | idct (double *v, size_t len) |
| double * | pad_to_power_of_two (double *v, size_t len, size_t *padded_len) |
| SFnc | idct_cf_to_sf (CFnc cf) |
| CFnc | dct_sf_to_cf (SFnc sf) |
| SFnc | desymmetrize_schofield_sf (SFnc sf) |
| Spectrum | desymmetrize_schofield_sp (Spectrum sp) |
| Spectrum | inv_desymmetrize_schofield (Spectrum sp) |
| SFnc | desymmetrize_d2_sf (SFnc sf) |
| Spectrum | desymmetrize_d2_sp (Spectrum sp) |
| Spectrum | inv_desymmetrize_d2 (Spectrum sp) |
| SFnc | desymmetrize_d1 (SFnc sf) |
| CFnc | egelstaff_time_transform (CFnc cf, bool frommhold_renormalization) |
| SFnc | desymmetrize_frommhold (SFnc sf) |
| SFnc | desymmetrize_egelstaff_from_cf (CFnc cf) |
| SFnc | desymmetrize_frommhold_from_cf (CFnc cf) |
| SFnc | desymmetrize_egelstaff (SFnc sf) |
| Spectrum | compute_alpha (SFnc sf) |
| double | integrate_composite_simpson (double *x, double *y, size_t len) |
| bool | compute_Mn_from_cf_using_classical_detailed_balance (CFnc cf, size_t n, double *result) |
| double | compute_Mn_from_sf_using_classical_detailed_balance (SFnc sf, size_t n) |
| double | compute_Mn_from_sf_using_quantum_detailed_balance (SFnc sf, size_t n) |
| CFnc | copy_cfnc (CFnc cf) |
| SFnc | copy_sfnc (SFnc sf) |
| Spectrum | copy_spectrum (Spectrum sp) |
Variables | |
| dipolePtr | dipole_1 = NULL |
| dipoleFree | free_dipole_1 = NULL |
| dipolePtr | dipole_2 = NULL |
| dipoleFree | free_dipole_2 = NULL |
| pesPtr | pes = NULL |
| dpesPtr | dpes = NULL |
| int | _wrank = 0 |
| int | _wsize = 1 |
| bool | _print0_suppress_info = false |
| int | _print0_margin = 0 |
| const char * | PAIR_STATES [PAIR_STATE_COUNT] |
| const char * | CALCULATION_TYPES [CALCULATION_TYPES_COUNT] |
| MonomerType | MONOMER_TYPES [MONOMER_COUNT] |
| WingParams | INIT_WP |
Macro Definition Documentation
◆ ARENA_IMPLEMENTATION
| #define ARENA_IMPLEMENTATION |
◆ ARENA_REGION_DEFAULT_CAPACITY
| #define ARENA_REGION_DEFAULT_CAPACITY (8*1024) |
◆ DEFAULT_JFIN_HISTOGRAM_BINS
| #define DEFAULT_JFIN_HISTOGRAM_BINS 352 |
◆ DEFAULT_JFIN_HISTOGRAM_FILENAME1
| #define DEFAULT_JFIN_HISTOGRAM_FILENAME1 "jfin.dat.1" |
◆ DEFAULT_JFIN_HISTOGRAM_FILENAME2
| #define DEFAULT_JFIN_HISTOGRAM_FILENAME2 "jfin.dat.2" |
◆ DEFAULT_JFIN_HISTOGRAM_MAX
| #define DEFAULT_JFIN_HISTOGRAM_MAX 35.0 |
◆ DEFAULT_JINI_HISTOGRAM_BINS
| #define DEFAULT_JINI_HISTOGRAM_BINS 352 |
◆ DEFAULT_JINI_HISTOGRAM_FILENAME1
| #define DEFAULT_JINI_HISTOGRAM_FILENAME1 "jini.dat.1" |
◆ DEFAULT_JINI_HISTOGRAM_FILENAME2
| #define DEFAULT_JINI_HISTOGRAM_FILENAME2 "jini.dat.2" |
◆ DEFAULT_JINI_HISTOGRAM_MAX
| #define DEFAULT_JINI_HISTOGRAM_MAX 35.0 |
◆ DEFAULT_NSWITCH_HISTOGRAM_BINS
| #define DEFAULT_NSWITCH_HISTOGRAM_BINS 20 |
◆ DEFAULT_NSWITCH_HISTOGRAM_FILENAME1
| #define DEFAULT_NSWITCH_HISTOGRAM_FILENAME1 "nswitch.dat.1" |
◆ DEFAULT_NSWITCH_HISTOGRAM_FILENAME2
| #define DEFAULT_NSWITCH_HISTOGRAM_FILENAME2 "nswitch.dat.2" |
◆ DEFAULT_NSWITCH_HISTOGRAM_MAX
| #define DEFAULT_NSWITCH_HISTOGRAM_MAX 20.0 |
◆ HISTOGRAM_MAX_TPS
| #define HISTOGRAM_MAX_TPS 50 |
◆ IMAG
| #define IMAG | ( | z, | |
| i ) |
◆ nparams
| #define nparams 3 |
◆ R_HISTOGRAM_BINS
| #define R_HISTOGRAM_BINS 100 |
◆ R_HISTOGRAM_MAX
| #define R_HISTOGRAM_MAX 10000000.0 |
◆ REAL
| #define REAL | ( | z, | |
| i ) |
Function Documentation
◆ analytic_full_partition_function_by_V()
| double analytic_full_partition_function_by_V | ( | MoleculeSystem * | ms, |
| double | Temperature ) |
◆ arena_linspace()
| double * arena_linspace | ( | Arena * | a, |
| double | start, | ||
| double | end, | ||
| size_t | n ) |
◆ arena_linspace_size_t()
| size_t * arena_linspace_size_t | ( | Arena * | a, |
| size_t | start, | ||
| size_t | end, | ||
| size_t | n ) |
◆ assert_float_is_equal_to()
| int assert_float_is_equal_to | ( | double | estimate, |
| double | true_value, | ||
| double | abs_tolerance ) |
◆ average_correlation_functions()
◆ average_correlation_functions__impl()
| int average_correlation_functions__impl | ( | CFnc * | average, |
| int | arg_count, | ||
| ... ) |
◆ blackman_apodization()
| void blackman_apodization | ( | Array | a, |
| double | sampling_time ) |
◆ calculate_correlation_and_save()
| CFnc calculate_correlation_and_save | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | Temperature ) |
calculate_correlation_and_save
TODO: check 'sampler_Rmin': we want to catch the situation when it's too low for given Temperature
◆ calculate_correlation_array_and_save()
| CFncArray calculate_correlation_array_and_save | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | base_temperature ) |
◆ calculate_M0()
| void calculate_M0 | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | Temperature, | ||
| double * | m, | ||
| double * | q ) |
calculate_M0 Running mean/variance formulas taken from GSL 1.15 https://github.com/ampl/gsl/blob/master/monte/plain.c
◆ calculate_M2()
| void calculate_M2 | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | Temperature, | ||
| double * | m, | ||
| double * | q ) |
calculate_M2 Running mean/variance formulas taken from GSL 1.15 https://github.com/ampl/gsl/blob/master/monte/plain.c
◆ calculate_spectral_function_using_prmu_representation_and_save()
| SFnc calculate_spectral_function_using_prmu_representation_and_save | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | Temperature ) |
◆ check_if_requantization_should_be_applied_to_monomer()
| void check_if_requantization_should_be_applied_to_monomer | ( | Monomer * | m, |
| size_t | step_counter ) |
◆ compute_alpha()
◆ compute_dHdp()
| void compute_dHdp | ( | MoleculeSystem * | ms, |
| gsl_matrix * | dHdp ) |
◆ compute_Mn_from_cf_using_classical_detailed_balance()
| bool compute_Mn_from_cf_using_classical_detailed_balance | ( | CFnc | cf, |
| size_t | n, | ||
| double * | result ) |
◆ compute_Mn_from_sf_using_classical_detailed_balance()
| double compute_Mn_from_sf_using_classical_detailed_balance | ( | SFnc | sf, |
| size_t | n ) |
◆ compute_Mn_from_sf_using_quantum_detailed_balance()
| double compute_Mn_from_sf_using_quantum_detailed_balance | ( | SFnc | sf, |
| size_t | n ) |
◆ compute_numerical_derivatives()
| Array compute_numerical_derivatives | ( | double(* | f )(double *q), |
| double * | q, | ||
| size_t | len, | ||
| size_t | order ) |
◆ compute_numerical_jac()
| gsl_matrix * compute_numerical_jac | ( | void(* | transform_angles )(double *qlab, double *qmol), |
| double * | qlab, | ||
| size_t | ninput_coordinates, | ||
| size_t | noutput_coordinates, | ||
| size_t | order ) |
◆ compute_numerical_rhs()
| Array compute_numerical_rhs | ( | MoleculeSystem * | ms, |
| size_t | order ) |
compute_numerical_rhs Computes the right-hand side of Hamilton's equations of motion using finite-difference formulas. The returned Array contains derivatives whose order corresponds to variable ordering employed in the MoleculeSystem structure.
- Parameters
-
ms Pointer to the MoleculeSystem for which to compute the derivatives. order Order of the finite-difference formula to employ for calculation.
◆ connes_apodization()
| void connes_apodization | ( | Array | a, |
| double | sampling_time ) |
◆ copy_cfnc()
◆ copy_sfnc()
◆ copy_spectrum()
◆ correlation_eval()
| int correlation_eval | ( | Arena * | a, |
| MoleculeSystem * | ms, | ||
| Trajectory * | traj, | ||
| CalcParams * | params, | ||
| double * | crln, | ||
| size_t * | tps ) |
◆ correlation_eval_zimmerman_trick()
| int correlation_eval_zimmerman_trick | ( | Arena * | a, |
| MoleculeSystem * | ms, | ||
| Trajectory * | traj, | ||
| CalcParams * | params, | ||
| double * | crln, | ||
| size_t * | tps ) |
◆ correlation_eval_zimmerman_trick_free_metastable()
| int correlation_eval_zimmerman_trick_free_metastable | ( | Arena * | a, |
| MoleculeSystem * | ms, | ||
| Trajectory * | traj, | ||
| CalcParams * | params, | ||
| double * | crln, | ||
| size_t * | tps, | ||
| double | Temperature ) |
◆ dct()
| double * dct | ( | double * | v, |
| size_t | len ) |
◆ dct_sf_to_cf()
◆ desymmetrize_d1()
◆ desymmetrize_d2_sf()
◆ desymmetrize_d2_sp()
◆ desymmetrize_egelstaff()
◆ desymmetrize_egelstaff_from_cf()
◆ desymmetrize_frommhold()
◆ desymmetrize_frommhold_from_cf()
◆ desymmetrize_schofield_sf()
◆ desymmetrize_schofield_sp()
◆ display_monomer_type()
| const char * display_monomer_type | ( | MonomerType | t | ) |
◆ egelstaff_time_transform()
◆ extract_dVdq_and_write_into_monomers()
| void extract_dVdq_and_write_into_monomers | ( | MoleculeSystem * | ms | ) |
◆ extract_q()
| void extract_q | ( | double * | qp, |
| double * | q, | ||
| size_t | QP_SIZE ) |
◆ extract_q_and_write_into_ms()
| void extract_q_and_write_into_ms | ( | MoleculeSystem * | ms | ) |
◆ find_closest_half_integer()
| double find_closest_half_integer | ( | double | j | ) |
requantization to: 0.0, 0.5, 1.5, 2.5, 3.5 ...
◆ find_closest_integer()
| double find_closest_integer | ( | double | j | ) |
requantization to: 0.0, 1.0, 2.0, 3.0 ...
◆ fit_baseline()
| WingParams fit_baseline | ( | CFnc * | cf, |
| size_t | EXT_RANGE_MIN ) |
◆ free_cfnc()
| void free_cfnc | ( | CFnc | cf | ) |
◆ free_cfnc_array()
| void free_cfnc_array | ( | CFncArray | ca | ) |
◆ free_ms()
| void free_ms | ( | MoleculeSystem * | ms | ) |
◆ free_sfnc()
| void free_sfnc | ( | SFnc | sf | ) |
◆ free_spectrum()
| void free_spectrum | ( | Spectrum | sp | ) |
◆ free_stored_histogram()
| void free_stored_histogram | ( | StoredHistogram * | sh | ) |
◆ generate_normal()
| double generate_normal | ( | double | sigma | ) |
◆ get_qp_from_ms()
| void get_qp_from_ms | ( | MoleculeSystem * | ms, |
| Array * | qp ) |
◆ gsl_fft_square()
| void gsl_fft_square | ( | double * | farr, |
| size_t | N ) |
◆ gsl_histogram_extend_right()
| gsl_histogram * gsl_histogram_extend_right | ( | gsl_histogram * | h, |
| size_t | add_bins ) |
◆ gsl_multifit_callback()
| void gsl_multifit_callback | ( | const size_t | iter, |
| void * | params, | ||
| const gsl_multifit_nlinear_workspace * | w ) |
◆ gsl_nonlinear_opt()
| void gsl_nonlinear_opt | ( | size_t | n, |
| double * | x, | ||
| double * | y, | ||
| WingParams * | wing_params ) |
◆ Hamiltonian()
| double Hamiltonian | ( | MoleculeSystem * | ms | ) |
Function Hamiltonian calls kinetic_energy function to compute kinetic energy, assembles a contiguous vector of coordinates via calling extract_q_and_write_into_ms and passes it to external pes function. The obtained values are added and returned.
◆ idct()
| double * idct | ( | double * | v, |
| size_t | len ) |
◆ idct_cf_to_sf()
◆ init_ms()
| MoleculeSystem init_ms | ( | double | mu, |
| MonomerType | t1, | ||
| MonomerType | t2, | ||
| double * | II1, | ||
| double * | II2, | ||
| size_t | seed ) |
Function init_ms prepares the MoleculeSystem based on the specified monomer types, sets up memory buffers and initializes random number generator.
- Parameters
-
mu Reduced mass of the molecule pair t1 Type of the first monomer t2 Type of the second monomer I1 Inertia tensor values for the first monomer. If the monomer is MonomerType::ATOM, no values will be read from the pointer, so NULL can be passed. Two and three values are expected for linear molecule (MonomerType::LINEAR_MOLECULE, MonomerType::LINEAR_MOLECULE_REQ_INTEGER and MonomerType::LINEAR_MOLECULE_REQ_HALFINTEGER) and rotor (MonomerType::ROTOR and MonomerType::ROTOR_REQUANTIZED_ROTATION). I2 Inertia tensor values for the second monomer. seed Seed to set up random number generator (Mersenne Twister). A unique seed will be produced if 0 value is passed.
◆ init_ms_from_monomers()
| MoleculeSystem init_ms_from_monomers | ( | double | mu, |
| Monomer * | m1, | ||
| Monomer * | m2, | ||
| size_t | seed ) |
Function init_ms_from_monomers prepares the MoleculeSystem based on the provided Monomer structs, sets up memory buffers and initializes random number generator.
◆ integrate_composite_simpson()
| double integrate_composite_simpson | ( | double * | x, |
| double * | y, | ||
| size_t | len ) |
◆ inv_desymmetrize_d2()
◆ inv_desymmetrize_schofield()
◆ invert_momenta()
| void invert_momenta | ( | MoleculeSystem * | ms | ) |
◆ j_monomer()
| void j_monomer | ( | Monomer * | m, |
| double | j[3] ) |
j_monomer computes the magnitude of angular momentum of passed-in monomer. Currently, implemented only for linear molecules.
![\[ [\textrm{linear molecule}]: j = \begin{bmatrix}
- p_\theta \sin \varphi - p_\varphi \cos \varphi / \tan \theta \\
p_\theta \cos \varphi - p_\varphi \sin \varphi / \tan \theta \\
p_\varphi
\end{bmatrix}
\]](form_0.png)
◆ kaiser_apodization()
| void kaiser_apodization | ( | Array | a, |
| double | sampling_time ) |
◆ kinetic_energy()
| double kinetic_energy | ( | MoleculeSystem * | ms | ) |
Function kinetic_energy computes the kinetic energy at the phase-point stored in MoleculeSystem.
The kinetic energy in the laboratory frame of reference is calculated in parts. The part corresponding to intermolecular degrees of freedom:
![\[ T_\textrm{int} = \frac{p_R^2}{2 \mu} + \frac{p_\Theta^2}{2 \mu R^2} + \frac{p_\Phi^2}{2\mu R^2 \sin^2 \Theta};
\]](form_2.png)
to linear molecule:
![\[ T_\textrm{lin} = \frac{p_\theta^2}{2 I} + \frac{p_\varphi^2}{2 I \sin^2 \theta};
\]](form_3.png)
and to rotor:
![\[ T_\textrm{rotor} = \frac{1}{2 I_1 \sin^2 \theta_2} \Big[ \left( p_\varphi - p_\psi \cos \theta \right) \sin \psi + p_\theta \sin \theta \cos \psi \Big]^2 \\
+ \frac{1}{2 I_2 \sin^2 \theta} \Big[ \left( p_\varphi - p_\psi \cos \theta \right) \cos \psi - p_\theta \sin \theta \sin \psi \Big]^2 \\
+ \frac{1}{2 I_3} \left( p_\psi \right)^2.
\]](form_4.png)
◆ linspace()
| double * linspace | ( | double | start, |
| double | end, | ||
| size_t | n ) |
◆ mpi_calculate_M0()
| void mpi_calculate_M0 | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | Temperature, | ||
| double * | m, | ||
| double * | q ) |
◆ mpi_calculate_M2()
| void mpi_calculate_M2 | ( | MoleculeSystem * | ms, |
| CalcParams * | params, | ||
| double | Temperature, | ||
| double * | m, | ||
| double * | q ) |
◆ normalize_cfnc()
| void normalize_cfnc | ( | CFnc * | cf | ) |
◆ p_generator()
| void p_generator | ( | MoleculeSystem * | ms, |
| double | Temperature ) |
Function p_generator samples momenta 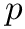
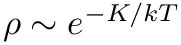
◆ p_generator_flux()
| void p_generator_flux | ( | MoleculeSystem * | ms, |
| double | Temperature ) |
Function p_generator samples momenta 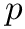
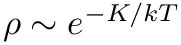
◆ pad_to_power_of_two()
| double * pad_to_power_of_two | ( | double * | v, |
| size_t | len, | ||
| size_t * | padded_len ) |
◆ parse_number_of_trajectories_from_header()
| bool parse_number_of_trajectories_from_header | ( | const char * | header, |
| double * | ntraj ) |
◆ parse_temperature_from_header()
| bool parse_temperature_from_header | ( | const char * | header, |
| double * | Temperature ) |
◆ put_qp_into_ms()
| void put_qp_into_ms | ( | MoleculeSystem * | ms, |
| Array | qp ) |
◆ q_generator()
| void q_generator | ( | MoleculeSystem * | ms, |
| CalcParams * | params ) |
Function q_generator generates 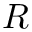
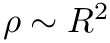
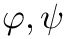
![$ \varphi, \psi \sim U[0, 2\pi] $](form_8.png)
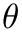
![$ \cos \theta \sim U[0, 1] $](form_10.png)
◆ read_correlation_function()
| bool read_correlation_function | ( | const char * | filename, |
| String_Builder * | sb, | ||
| CFnc * | cf ) |
◆ read_spectral_function()
| bool read_spectral_function | ( | const char * | filename, |
| String_Builder * | sb, | ||
| SFnc * | sf ) |
◆ read_spectrum()
| bool read_spectrum | ( | const char * | filename, |
| String_Builder * | sb, | ||
| Spectrum * | sp ) |
◆ recv_histogram_and_append()
| void recv_histogram_and_append | ( | Arena * | a, |
| int | source, | ||
| gsl_histogram ** | h ) |
◆ reject()
| bool reject | ( | MoleculeSystem * | ms, |
| double | Temperature, | ||
| double | pesmin ) |
reject applies the rejection step to the phase-point that is stored in the MoleculeSystem. It presupposes that the provided phase-point is sampled from 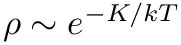
![$ u \sim U[0, 1] $](form_13.png)
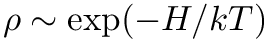
◆ rhs()
| int rhs | ( | realtype | t, |
| N_Vector | y, | ||
| N_Vector | ydot, | ||
| void * | data ) |
rhs function is passed to CVode library to propagate the trajectory. First, the phase-point coordinates are stored into MoleculeSystem struct. A contiguous vector of coordinates is assembled via extract_q_and_write_into_ms. Next, by calling the external function dpes, the derivatives of potential energy are computed and stored into MoleculeSystem::dVdq field. The components of derivative vector are then copied into the field Monomer::dVdq of the corresponding Monomer via the call to extract_dVdq_and_write_into_monomers. The right-hand side of Hamilton's equations with respect to intermolecular degrees of freedom are readily obtained and filled into output ydot, while the derivatives with respect to monomer's coordinates are handled by rhsMonomer function.
◆ rhsMonomer()
| void rhsMonomer | ( | Monomer * | m, |
| double * | deriv ) |
rhsMonomer dispatches between computing the right-hand side of Hamilton's equations of motion depending on the type of monomer. In addition to differentiating the kinetic energy, the derivatives of potential energy, which are taken from Monomer::dVdq, are also added to compute the right-hand side.
rhsMonomer does not account for requantization which should happen after this function has been called.
◆ sb_append()
| void sb_append | ( | String_Builder * | sb, |
| const char * | line, | ||
| size_t | n ) |
◆ sb_append_cstring()
| void sb_append_cstring | ( | String_Builder * | sb, |
| const char * | line ) |
◆ sb_append_format()
| void sb_append_format | ( | String_Builder * | sb, |
| const char * | format, | ||
| ... ) |
◆ sb_append_null()
| void sb_append_null | ( | String_Builder * | sb | ) |
◆ sb_append_seconds_as_datetime_string()
| void sb_append_seconds_as_datetime_string | ( | String_Builder * | sb, |
| int | s ) |
◆ sb_free()
| void sb_free | ( | String_Builder * | sb | ) |
◆ sb_reserve()
| void sb_reserve | ( | String_Builder * | sb, |
| size_t | n ) |
◆ sb_reset()
| void sb_reset | ( | String_Builder * | sb | ) |
◆ send_histogram_and_reset()
| void send_histogram_and_reset | ( | gsl_histogram * | h | ) |
◆ setup_jfin_histogram_for_monomer()
| void setup_jfin_histogram_for_monomer | ( | Monomer * | m | ) |
◆ setup_jini_histogram_for_monomer()
| void setup_jini_histogram_for_monomer | ( | Monomer * | m | ) |
◆ setup_nswitch_histogram_for_monomer()
| void setup_nswitch_histogram_for_monomer | ( | Monomer * | m | ) |
◆ torque_monomer()
| double torque_monomer | ( | Monomer * | m | ) |
torque_monomer computes the magnitude of torque (time-derivative of angular momentum) of passed-in monomer. Requires the derivative of potential Monomer::dVdq to be set within the monomer. Currently, implemented only for linear molecules.
![\[ [\textrm{linear molecule}]: \tau = \begin{bmatrix}
\displaystyle \sin \varphi \frac{\diff{V}}{\diff{\theta}} + \cos \varphi / \tan \theta \frac{\diff{V}}{\diff{\varphi}} \\
\displaystyle -\cos \varphi \frac{\diff{V}}{\diff{\theta}} + \sin \varphi / \tan \theta \frac{\diff{V}}{\diff{\varphi}} \\
\displaystyle -\frac{\diff{V}}{\diff{\varphi}}
\end{bmatrix}
\]](form_1.png)
◆ track_turning_points()
| void track_turning_points | ( | Tracker * | tr, |
| double | R ) |
◆ wingmodel()
| double wingmodel | ( | WingParams * | wp, |
| double | t ) |
◆ wingmodel_df()
| int wingmodel_df | ( | const gsl_vector * | x, |
| void * | data, | ||
| gsl_matrix * | J ) |
◆ wingmodel_f()
| int wingmodel_f | ( | const gsl_vector * | x, |
| void * | data, | ||
| gsl_vector * | f ) |
◆ wingmodel_image()
| double wingmodel_image | ( | WingParams * | wp, |
| double | nu ) |
◆ write_correlation_function()
| bool write_correlation_function | ( | const char * | filename, |
| CFnc | cf ) |
◆ write_correlation_function_ext()
| int write_correlation_function_ext | ( | FILE * | fp, |
| CFnc | cf ) |
◆ write_histogram_ext()
| int write_histogram_ext | ( | FILE * | fp, |
| gsl_histogram * | h, | ||
| int | count ) |
◆ write_spectral_function()
| bool write_spectral_function | ( | const char * | filename, |
| SFnc | sf ) |
◆ write_spectral_function_ext()
| int write_spectral_function_ext | ( | FILE * | fp, |
| SFnc | sf ) |
◆ write_spectrum()
| bool write_spectrum | ( | const char * | filename, |
| Spectrum | sp ) |
◆ write_spectrum_ext()
| int write_spectrum_ext | ( | FILE * | fp, |
| Spectrum | sp ) |
◆ writetxt()
| bool writetxt | ( | const char * | filename, |
| double * | x, | ||
| double * | y, | ||
| size_t | len, | ||
| const char * | header ) |
Variable Documentation
◆ _print0_margin
| int _print0_margin = 0 |
◆ _print0_suppress_info
| bool _print0_suppress_info = false |
◆ _wrank
| int _wrank = 0 |
◆ _wsize
| int _wsize = 1 |
◆ CALCULATION_TYPES
| const char* CALCULATION_TYPES[CALCULATION_TYPES_COUNT] |
◆ dipole_1
| dipolePtr dipole_1 = NULL |
◆ dipole_2
| dipolePtr dipole_2 = NULL |
◆ dpes
| dpesPtr dpes = NULL |
◆ free_dipole_1
| dipoleFree free_dipole_1 = NULL |
◆ free_dipole_2
| dipoleFree free_dipole_2 = NULL |
◆ INIT_WP
| WingParams INIT_WP |
◆ MONOMER_TYPES
| MonomerType MONOMER_TYPES[MONOMER_COUNT] |
◆ PAIR_STATES
| const char* PAIR_STATES[PAIR_STATE_COUNT] |
◆ pes
| pesPtr pes = NULL |
Generated by