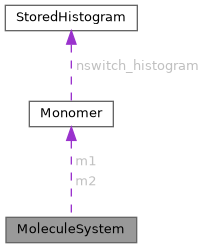
Angular variables and conjugated momenta are stored within the MoleculeSystem struct in the same order as within the qp of Monomer struct. More...
#include <hawaii.h>
Public Attributes | |
| double | intermolecular_qp [6] |
| Coordinates and conjugated momenta that correspond to the intermolecular motion ( | |
| Monomer | m1 |
| First monomer. | |
| Monomer | m2 |
| Second monomer. | |
| double | mu |
| Reduced mass of molecule pair. | |
| size_t | Q_SIZE |
| Total number of coordinates for molecule pair. | |
| size_t | QP_SIZE |
| Total number of coordinates and momenta for molecular pair (phase point size). | |
| double * | intermediate_q |
| Contiguous vector of coordinates. | |
| double * | dVdq |
| Contiguous vector of potential energy derivatives. | |
| size_t | seed |
| time_t | init_rawtime |
| Initialization time represented as the number of seconds since Unix Epoch; set in init_ms. | |
| time_t | temp_rawtime |
| A temporary time field for the time at which the previous iteration of the iterative algorithm was completed; used for tracking elapsed time between iterations. | |
Detailed Description
Angular variables and conjugated momenta are stored within the MoleculeSystem struct in the same order as within the qp of Monomer struct.
The variables' indices in the intermolecular_qp array are defined as follows:
Keep in mind that intermolecular coordinates and monomer's coordinates are not stored contiguously (monomer's coordinates are stored within the Monomer structure). The contiguous vector of coordinates can be assembled by calling extract_q_and_write_into_ms function, which stores the coordinates in memory pointed at by intermediate_q. These coordinates are passed to external functions that compute the values of intermolecular energy, its derivatives with respect to coordinates and induced dipole. There is no guarantee that coordinates stored in Monomer's and coordinates in memory at intermediate_q are always in sync. The function extract_q_and_write_into_ms must be invoked if the contiguous vector of coordinates is desired at a certain point of the program execution.
Member Data Documentation
◆ dVdq
| double* MoleculeSystem::dVdq |
Contiguous vector of potential energy derivatives.
◆ init_rawtime
| time_t MoleculeSystem::init_rawtime |
Initialization time represented as the number of seconds since Unix Epoch; set in init_ms.
◆ intermediate_q
| double* MoleculeSystem::intermediate_q |
Contiguous vector of coordinates.
◆ intermolecular_qp
| double MoleculeSystem::intermolecular_qp[6] |
Coordinates and conjugated momenta that correspond to the intermolecular motion ( 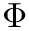
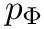

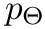
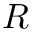
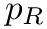
◆ m1
| Monomer MoleculeSystem::m1 |
First monomer.
◆ m2
| Monomer MoleculeSystem::m2 |
Second monomer.
◆ mu
| double MoleculeSystem::mu |
Reduced mass of molecule pair.
◆ Q_SIZE
| size_t MoleculeSystem::Q_SIZE |
Total number of coordinates for molecule pair.
◆ QP_SIZE
| size_t MoleculeSystem::QP_SIZE |
Total number of coordinates and momenta for molecular pair (phase point size).
◆ seed
| size_t MoleculeSystem::seed |
◆ temp_rawtime
| time_t MoleculeSystem::temp_rawtime |
A temporary time field for the time at which the previous iteration of the iterative algorithm was completed; used for tracking elapsed time between iterations.
The documentation for this struct was generated from the following file:
- src/hawaii.h
Generated by